2019
University of Chicago, Center for Translational Data Science
Founding Product Designer & Researcher
Gen3 Data Commons is a tool that is supported by the National Institutes of Health (NIH) to store data from research projects that are sponsored by NIH. However, the biometrics files are usually very large and take a long time to upload, so they are often sent through the mail on flash drives.(Yes, through Fedex!) This makes it difficult to connect the files to their related medical information and also makes it difficult for NIH to store and reuse the data.
We wanted to create a tool that makes it easy, fast, and safe to upload large amounts of data files digitally, and also allows for these files to be stored with the correct medical information in the Data Commons.
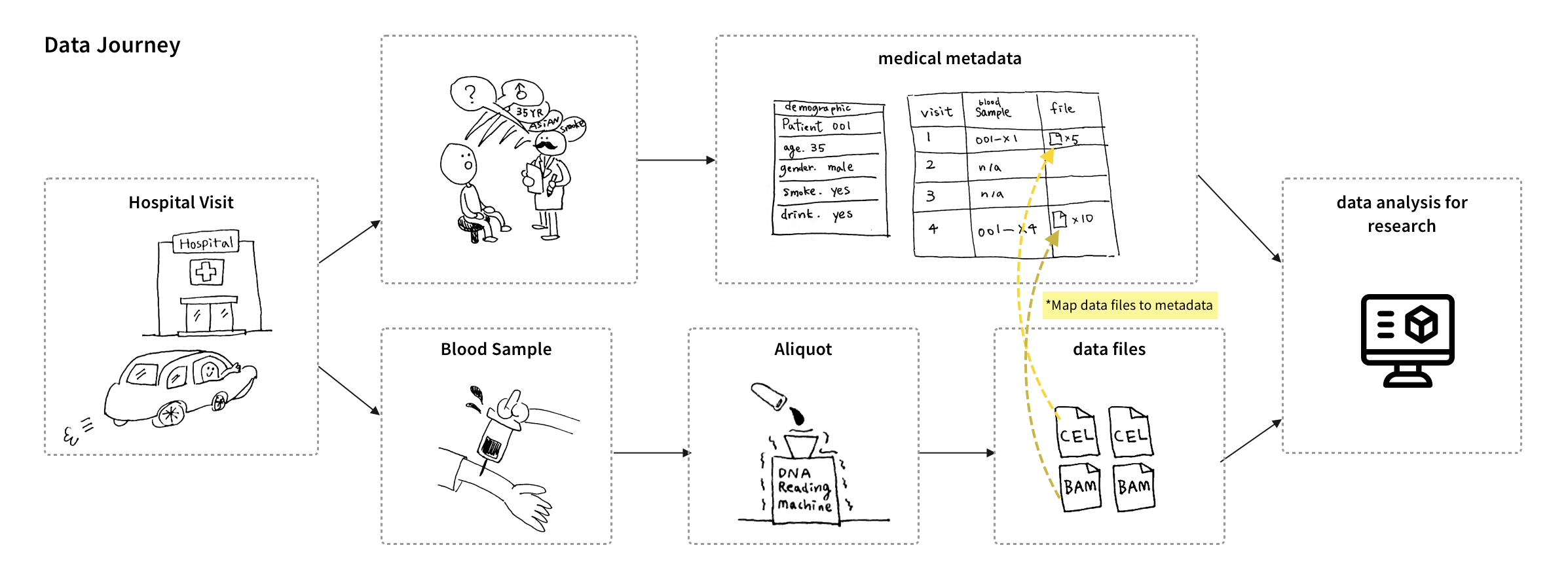
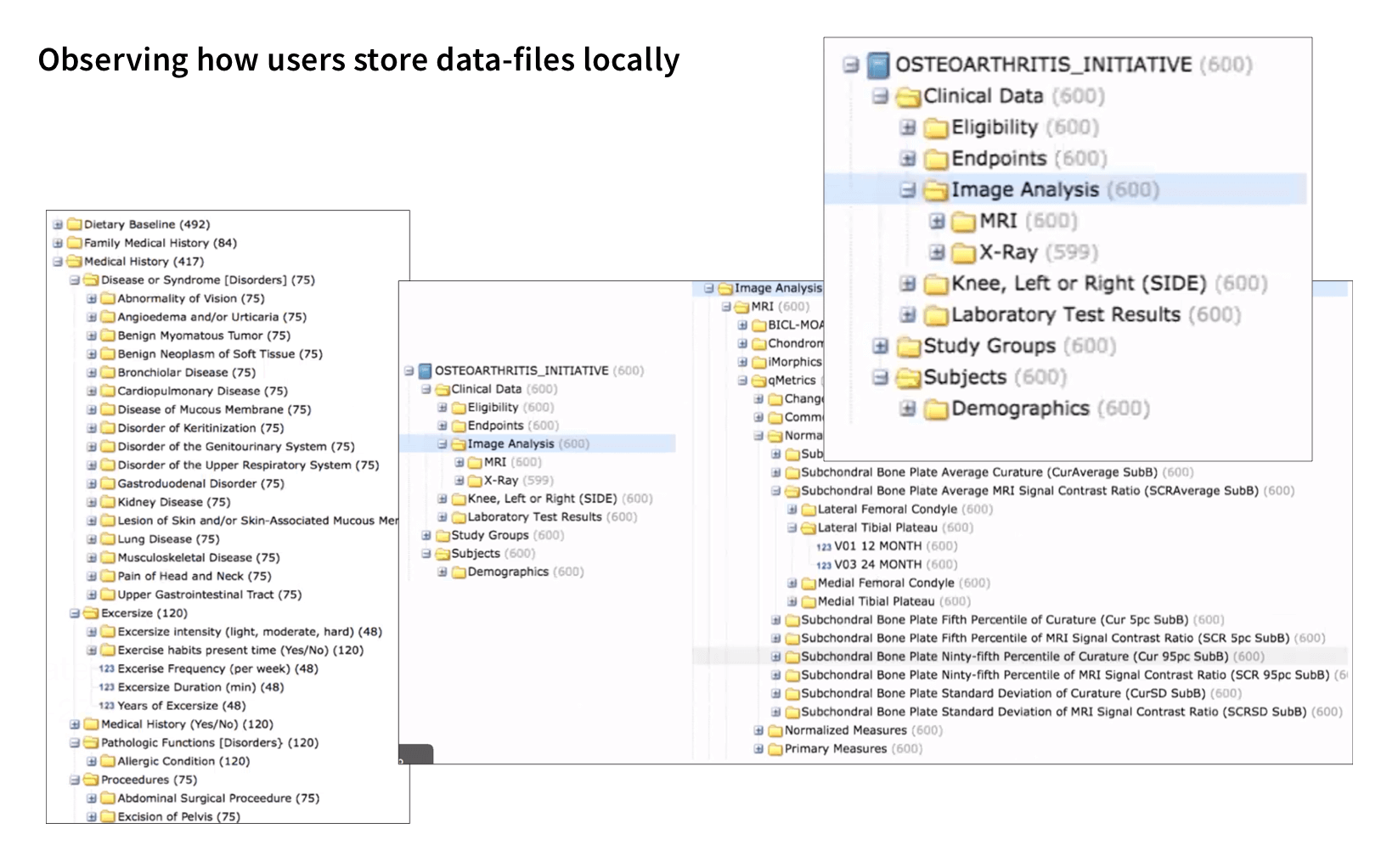
I visited hospitals and labs to understand how biometrics files get collected, used, and stored. During the visits, I got to see how each data type has its unique process to be collected and stored. For MVP, we worked with the stakeholders to prioritize 2 types of data files:
Genome DNA sequencing data
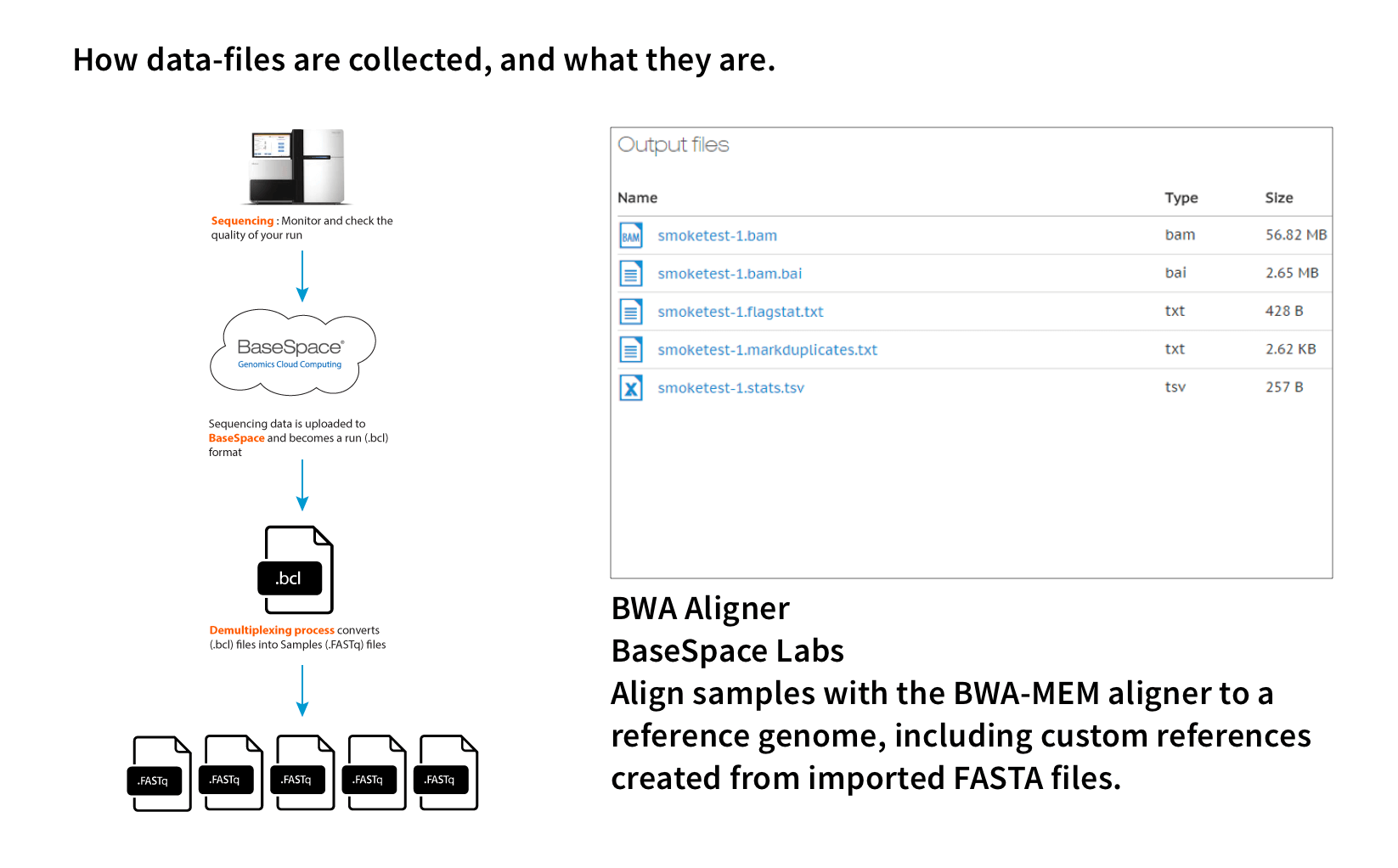
The data is usually in FASTQ or BAM format, and each file is about 90 GB. For a whole study, it can be between 270 GB and 3 TB.
MRI image files
The data is usually in JPG or PNG format, and each file is about 50 MB. For a whole study, it can include 9,000+ images.
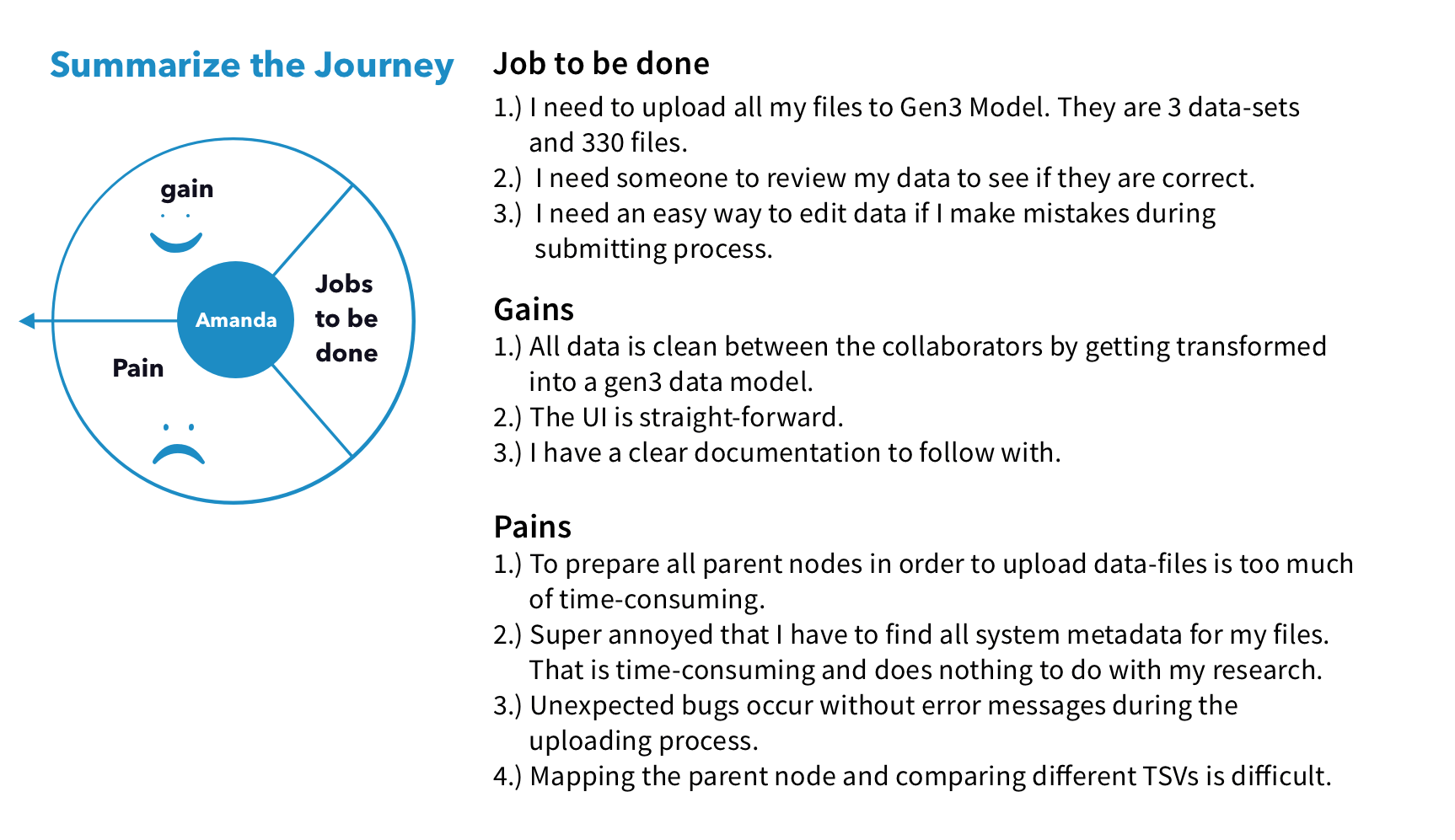
I conducted interviews with users who work with our targeted data types, specifically in the areas of Cancer, TB, and Brain research. During these interviews, I observed users' data files, analyzed their workflow, and gained insight into the tools they use at each step. After that, I created personas and journey maps to share the findings with the team.
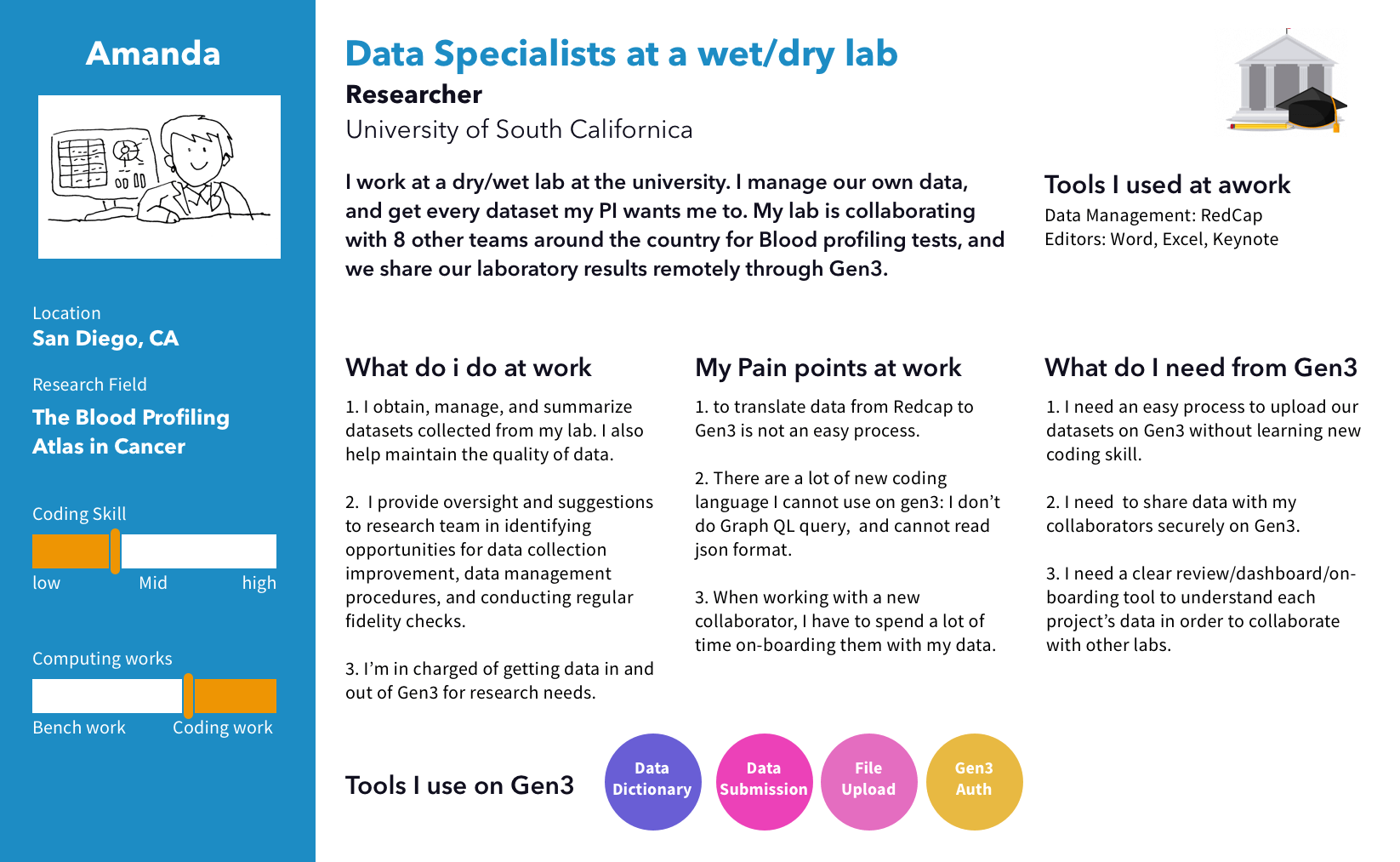
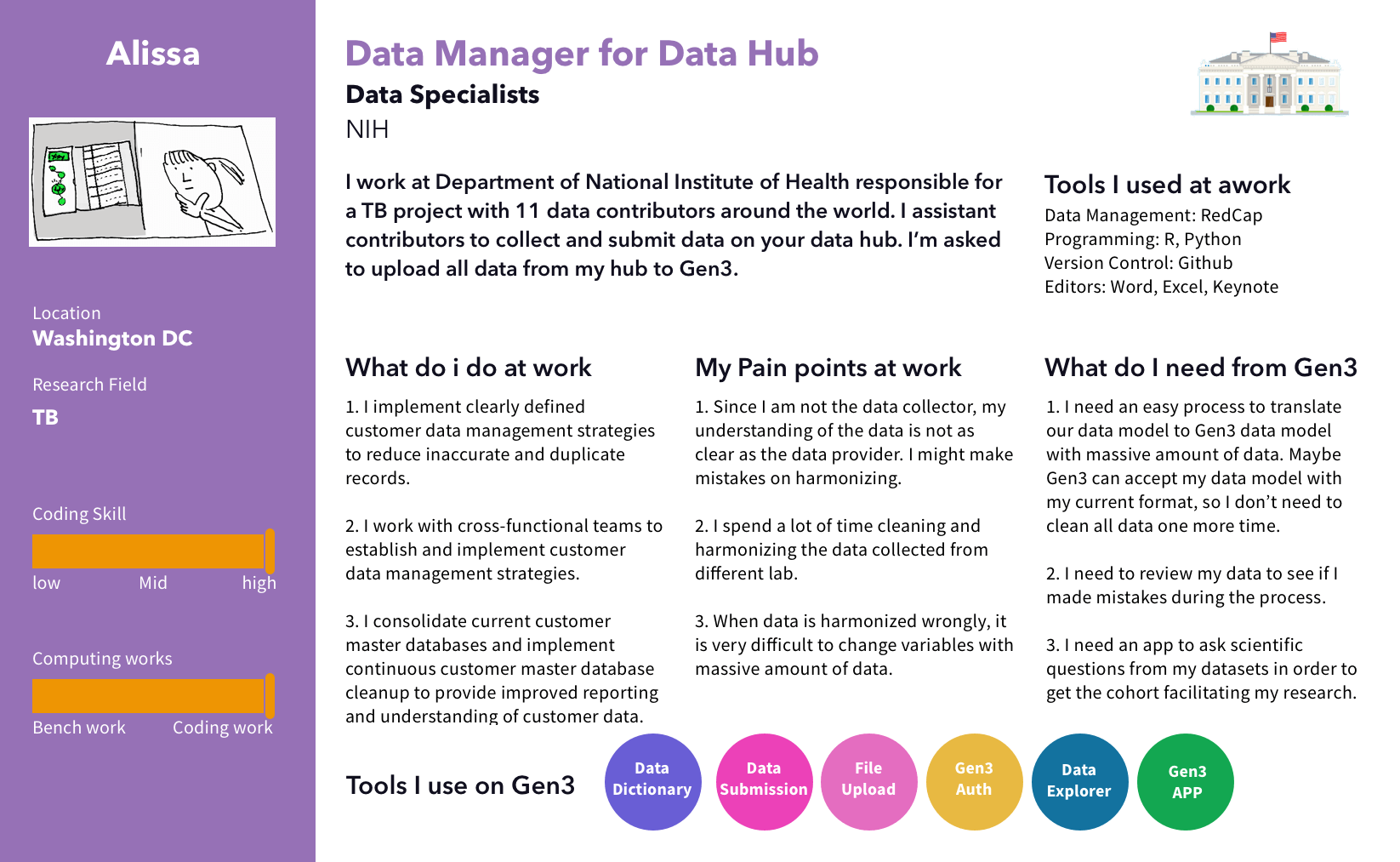
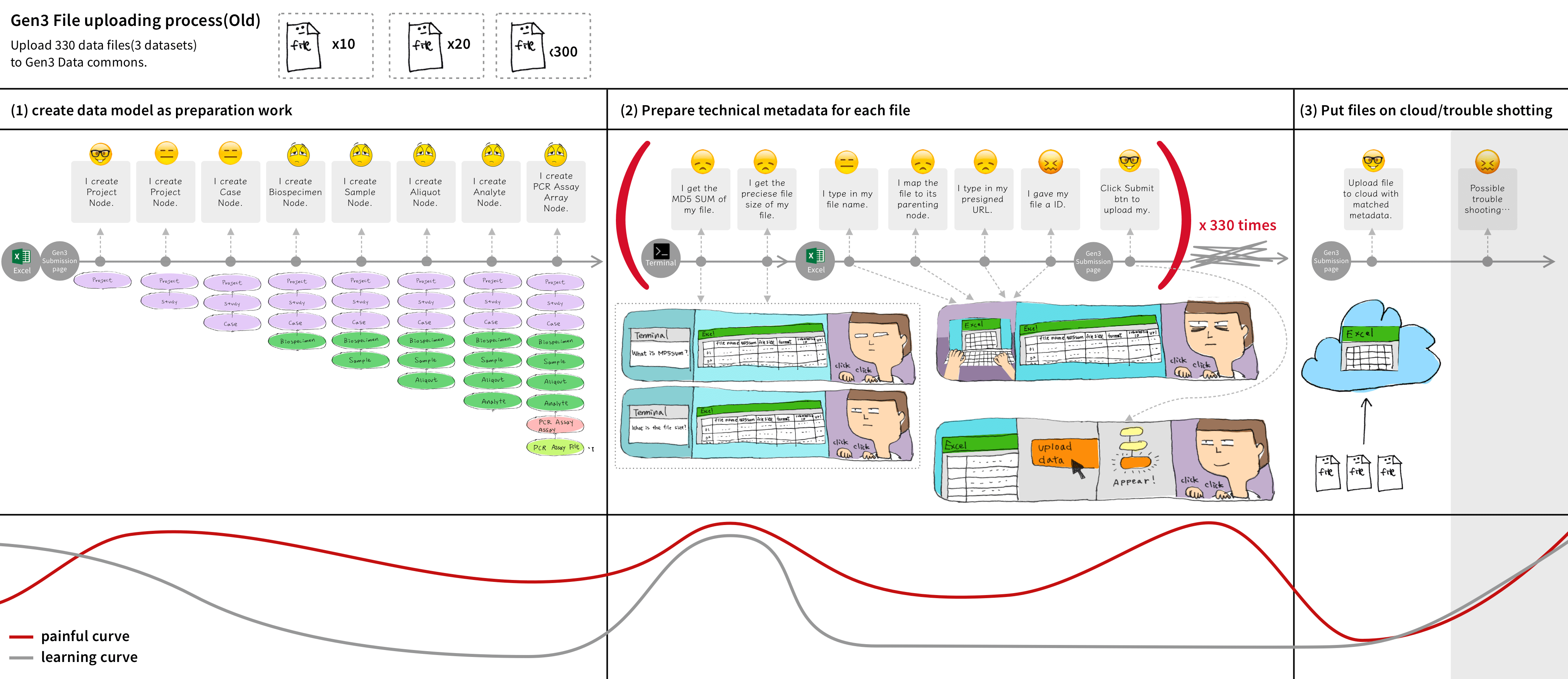
We discovered 2 main pain points for uploading files. First, uploading large data files on a web browser failed a lot due to session timeouts. Second, users cannot upload files before the time-consuming process of creating nodes to define the file's metadata structure.
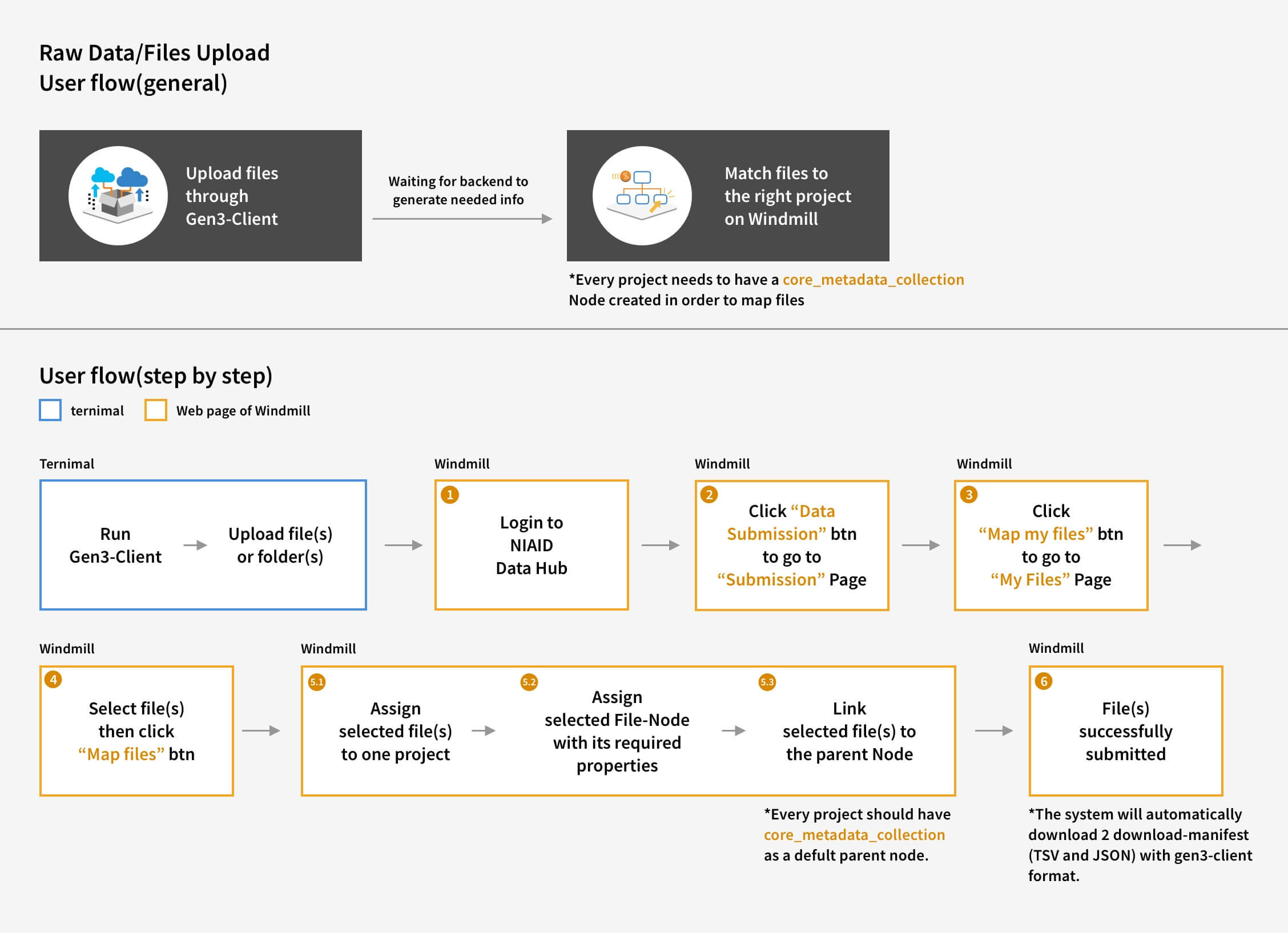
We made Gen3-Client based on GO to upload files through command lines in Terminal, allowing users to have a stable environment uploading files. Also, we built a web UI for users to assign upload files to a project, skipping the complicated medical metadata mapping.
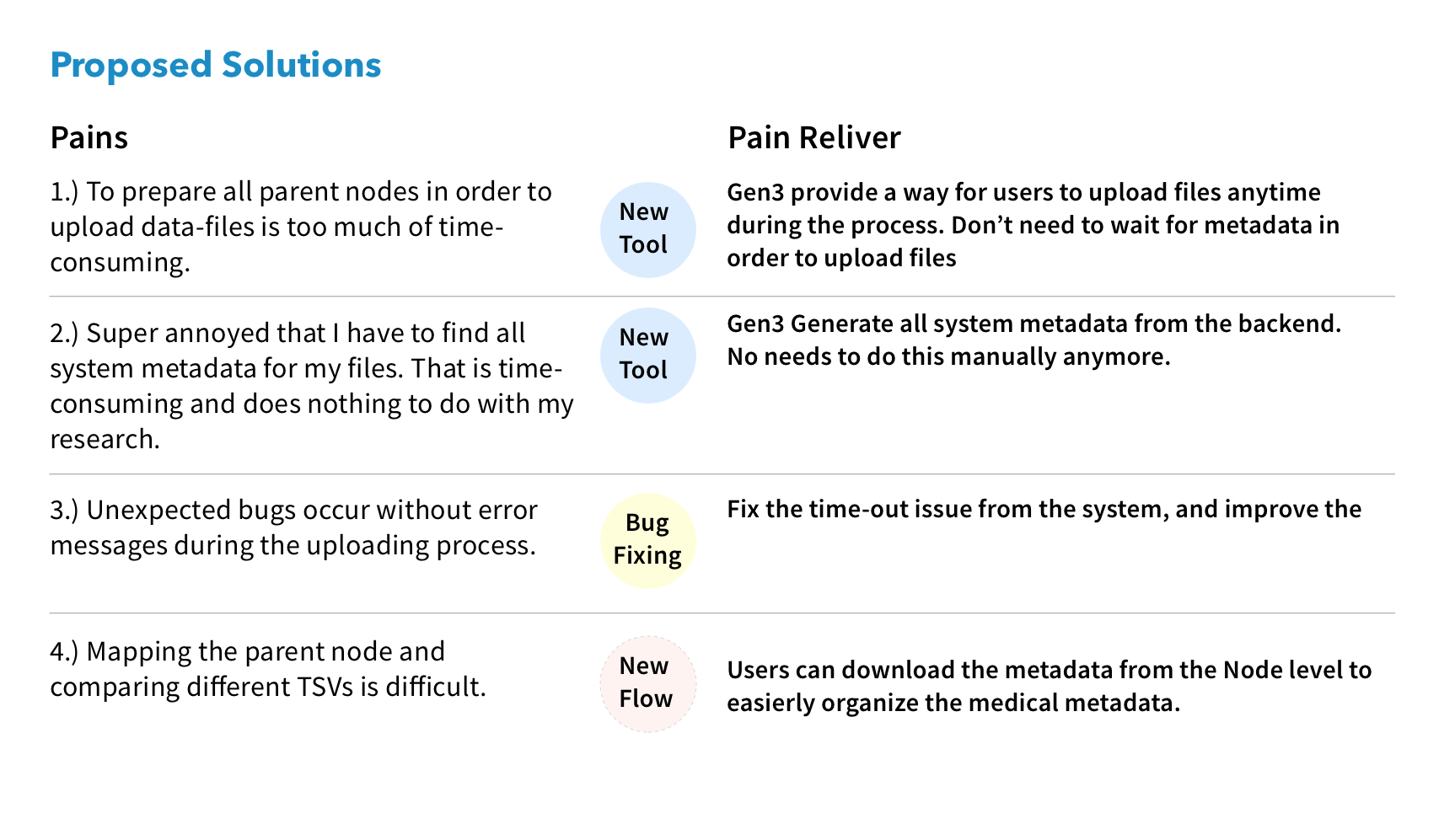
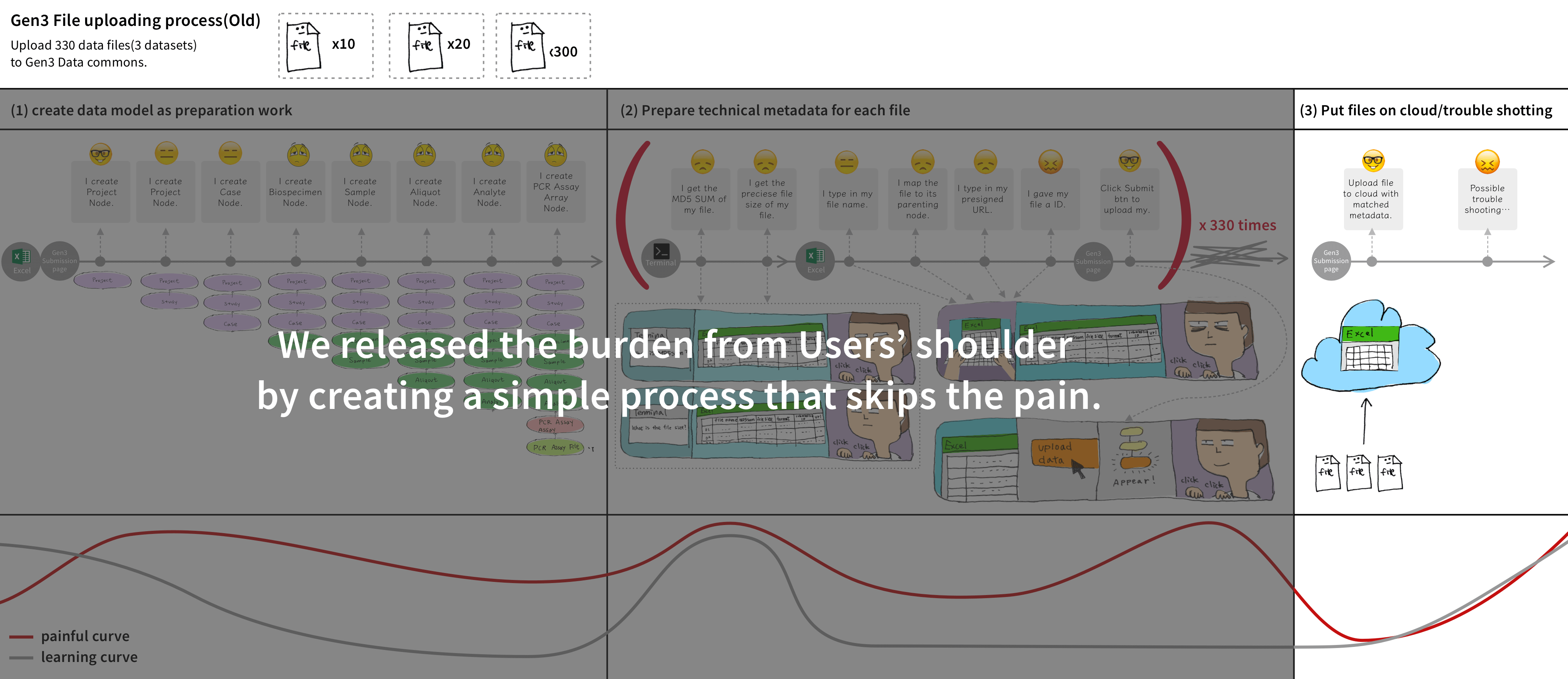
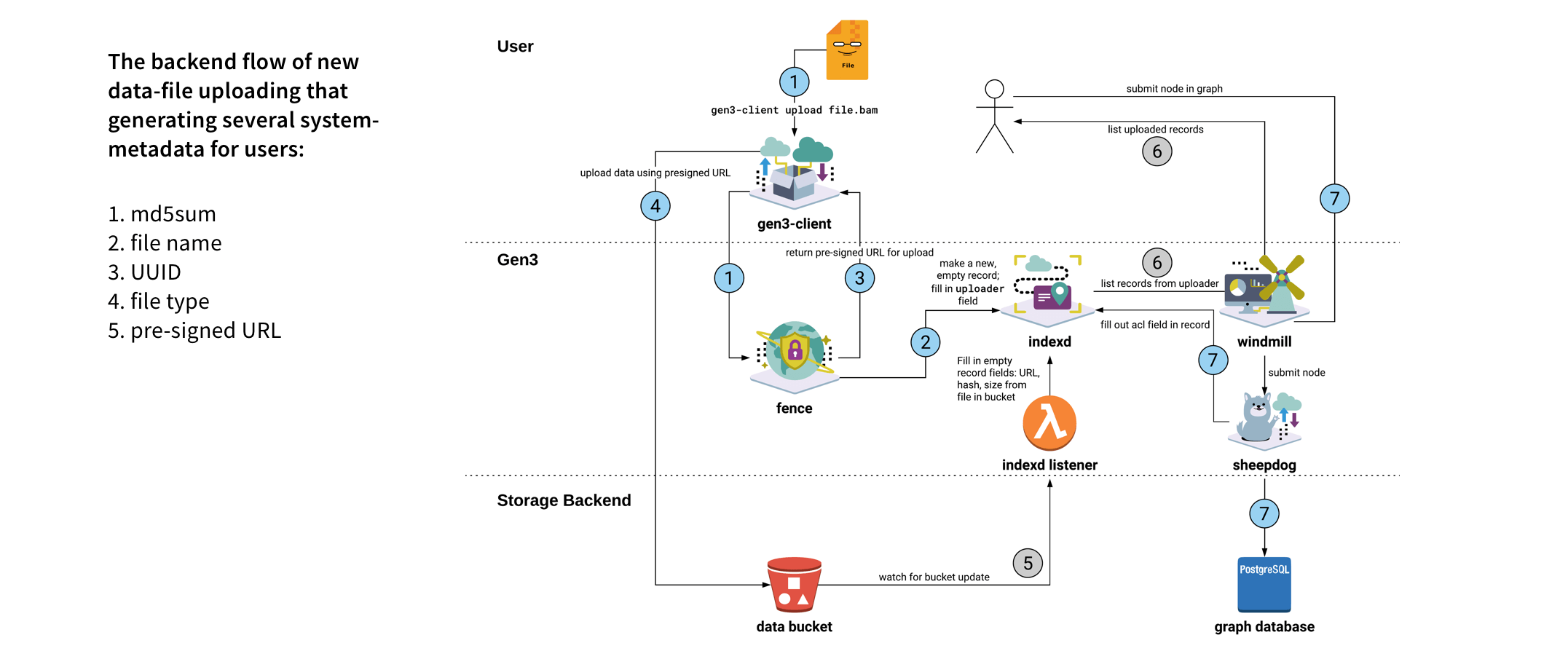
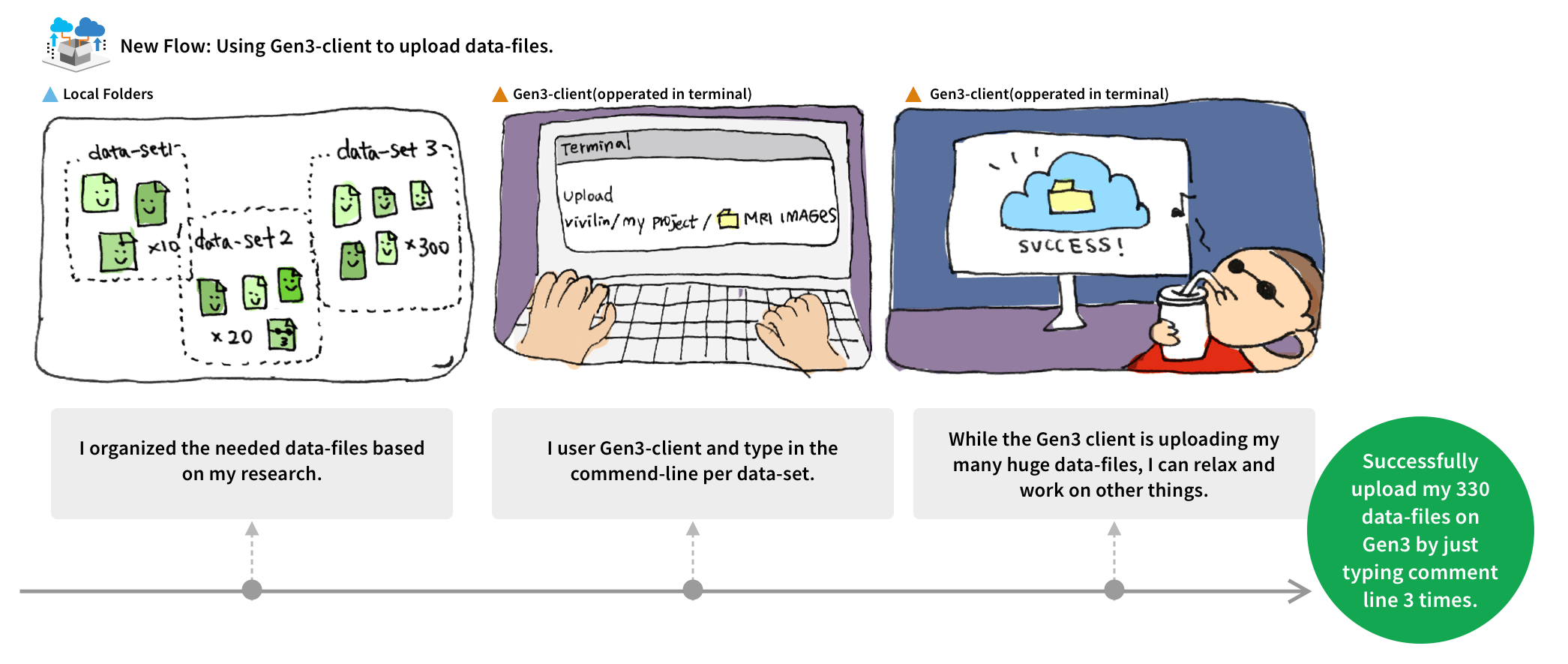
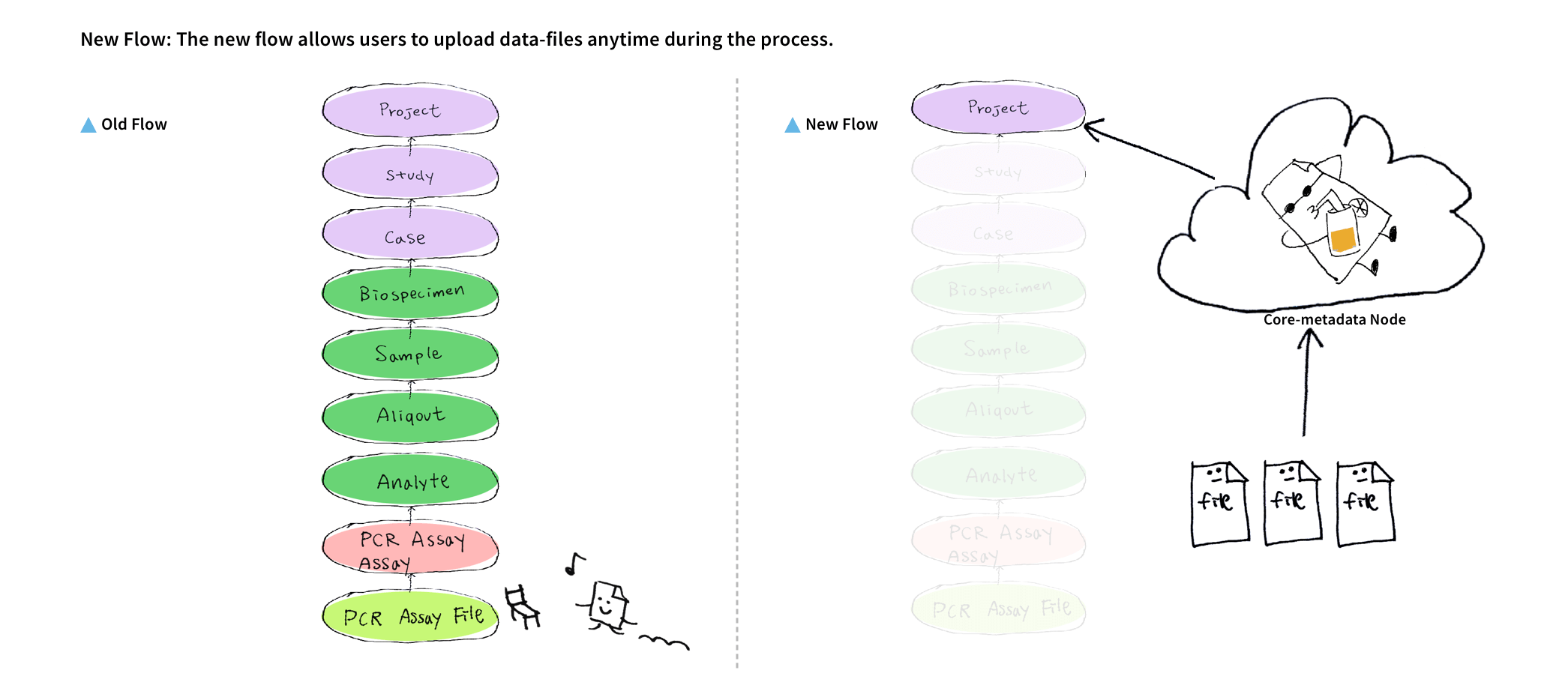
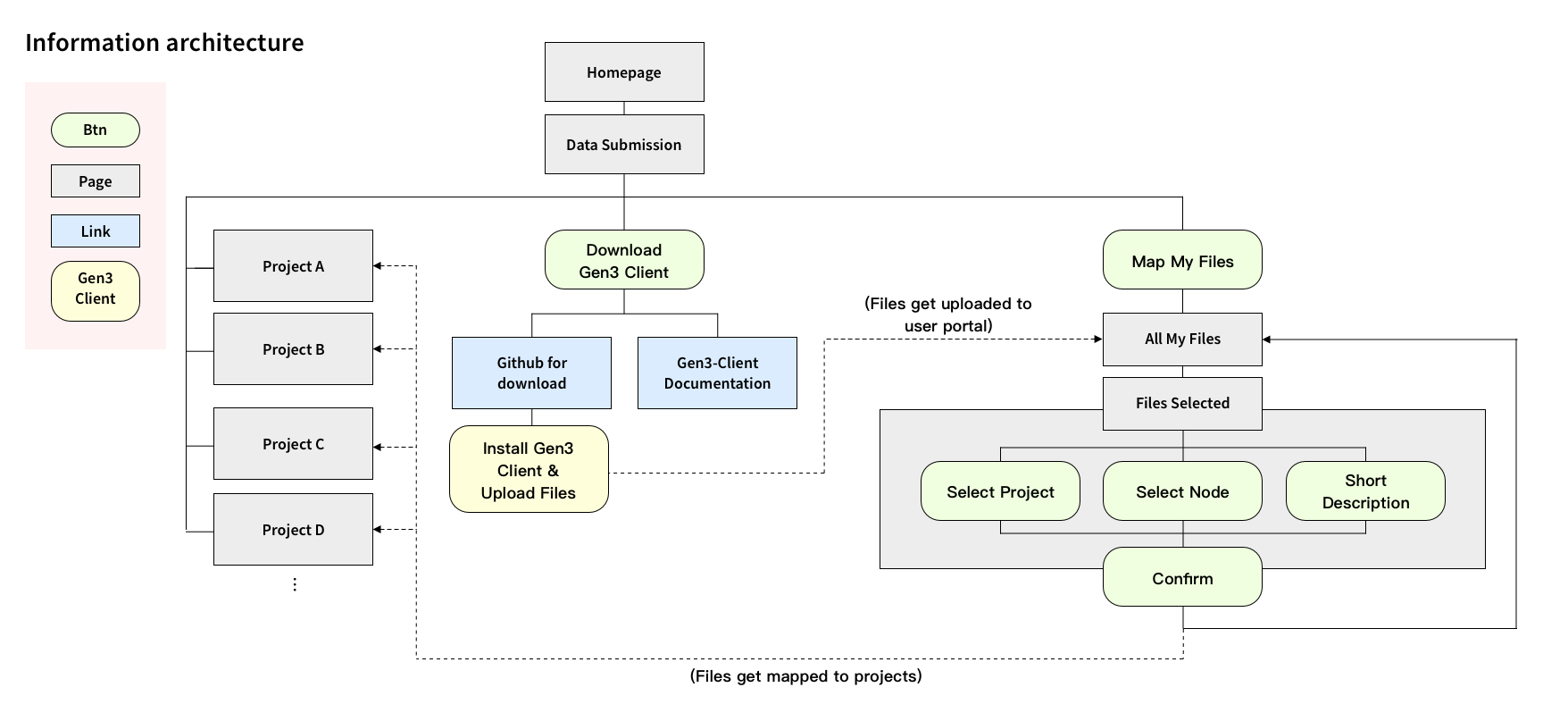
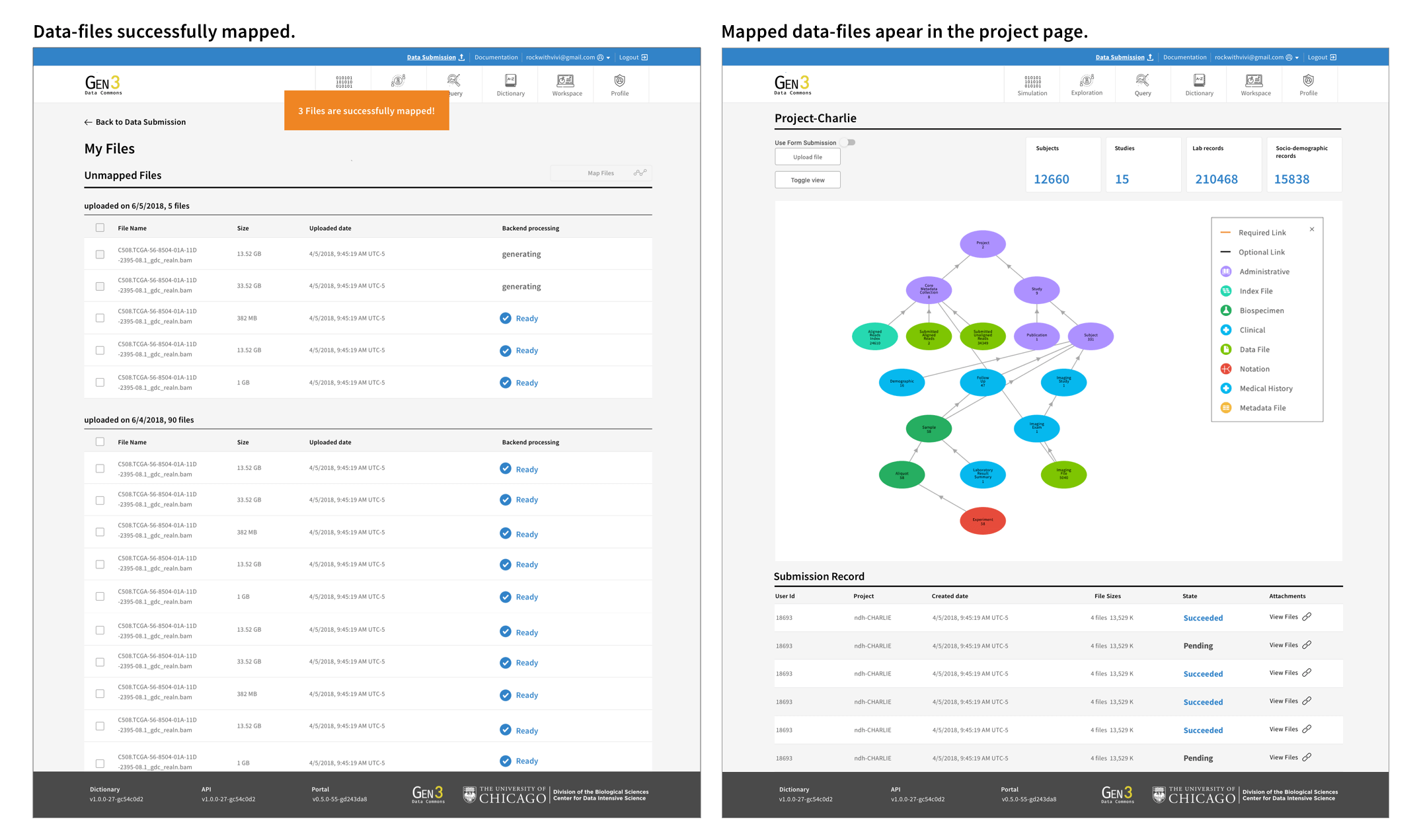
I reorganized the information architecture to implement a new workflow for users, which involved downloading a file-uploading tool to upload files, and then returning to the user interface to map the uploaded files with their related medical metadata.
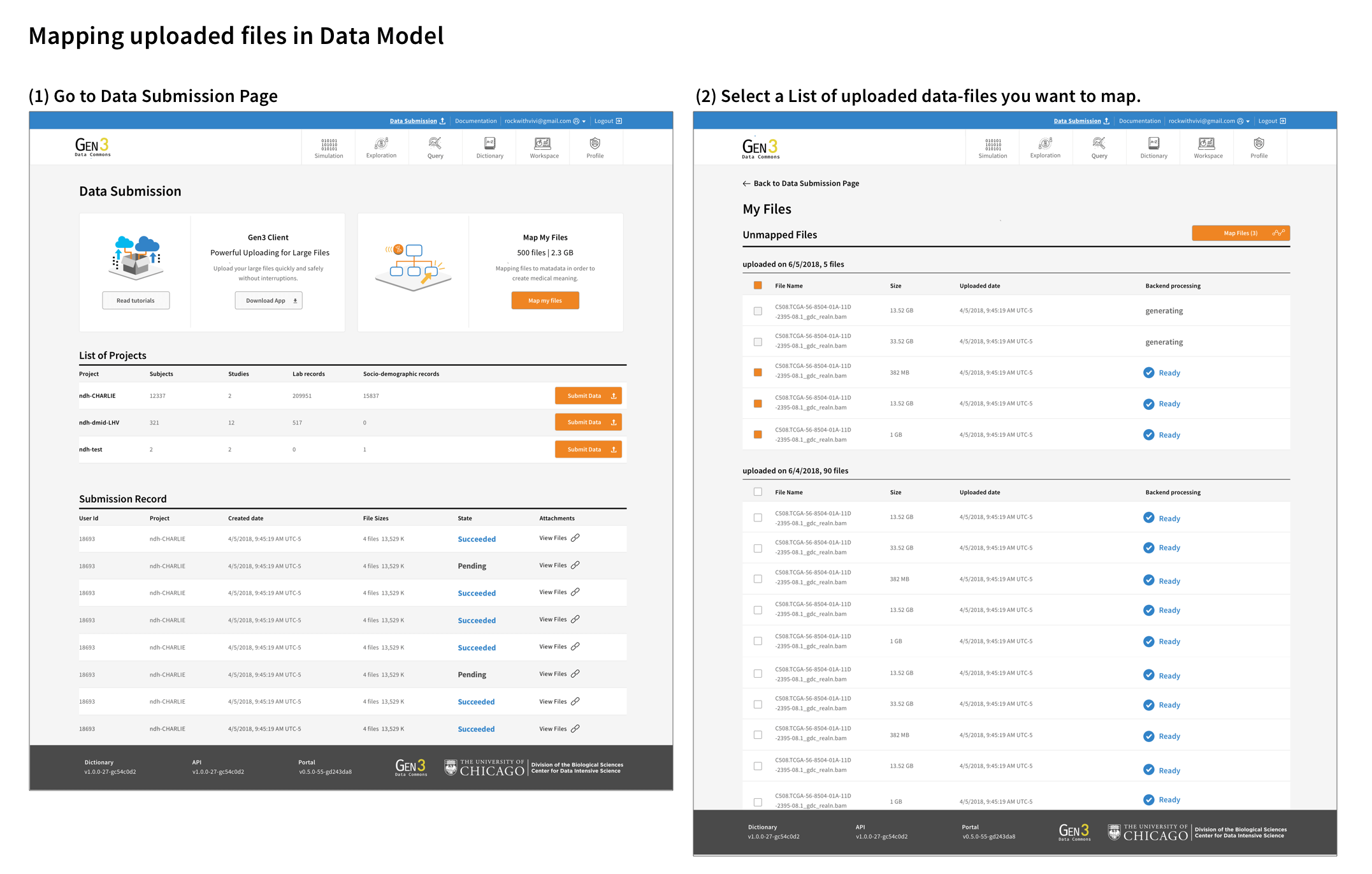
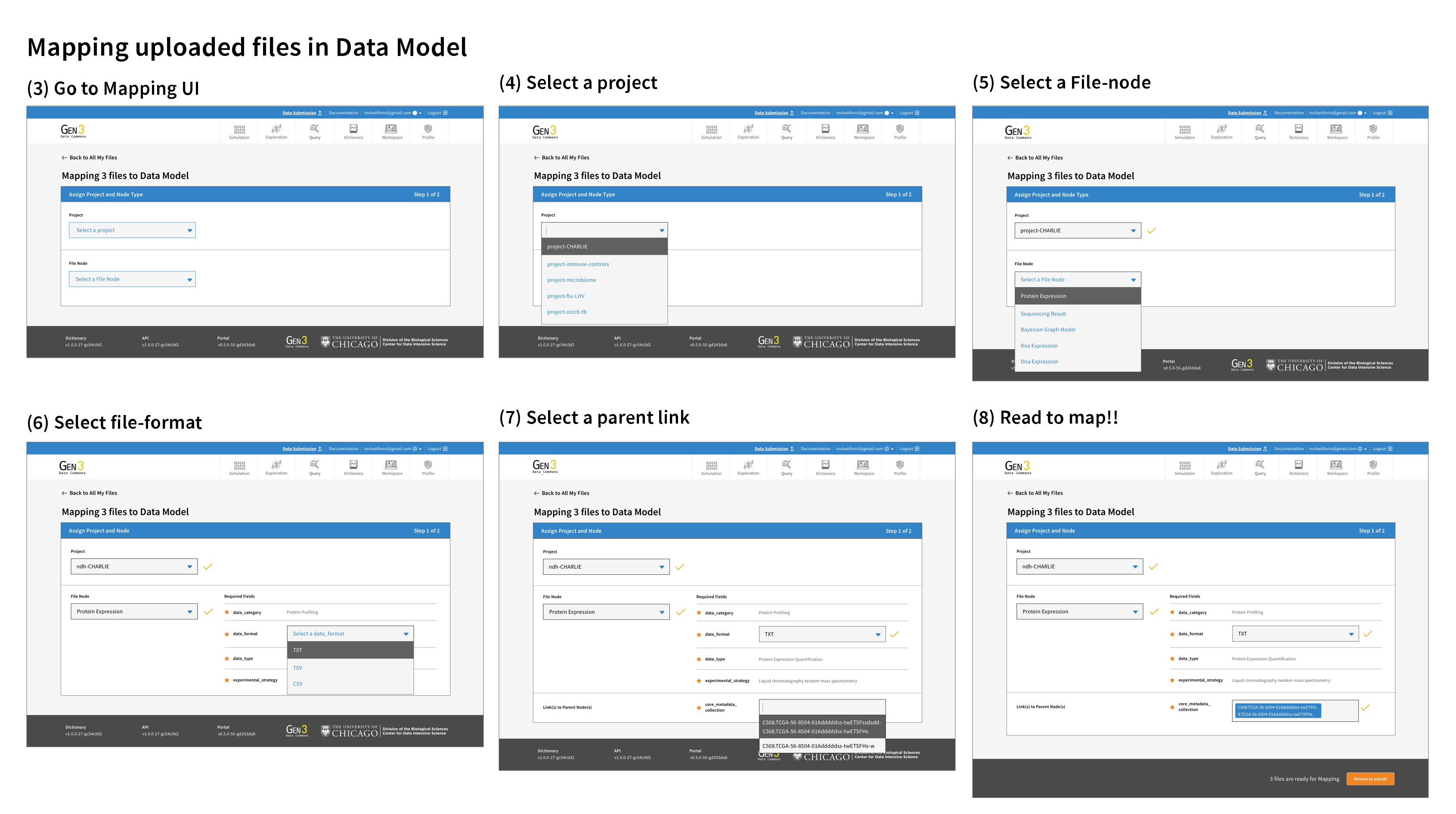
The goal of the metadata mapping UI design was to be simple, intuitive, and easy to navigate. One of the main interactive challenges was to effectively convey the various file uploading statuses and organized the unmapped files. I made it necessary for users to connect the files to a project and also added the ability for users to add notes to the files so they can come back to organize files later.
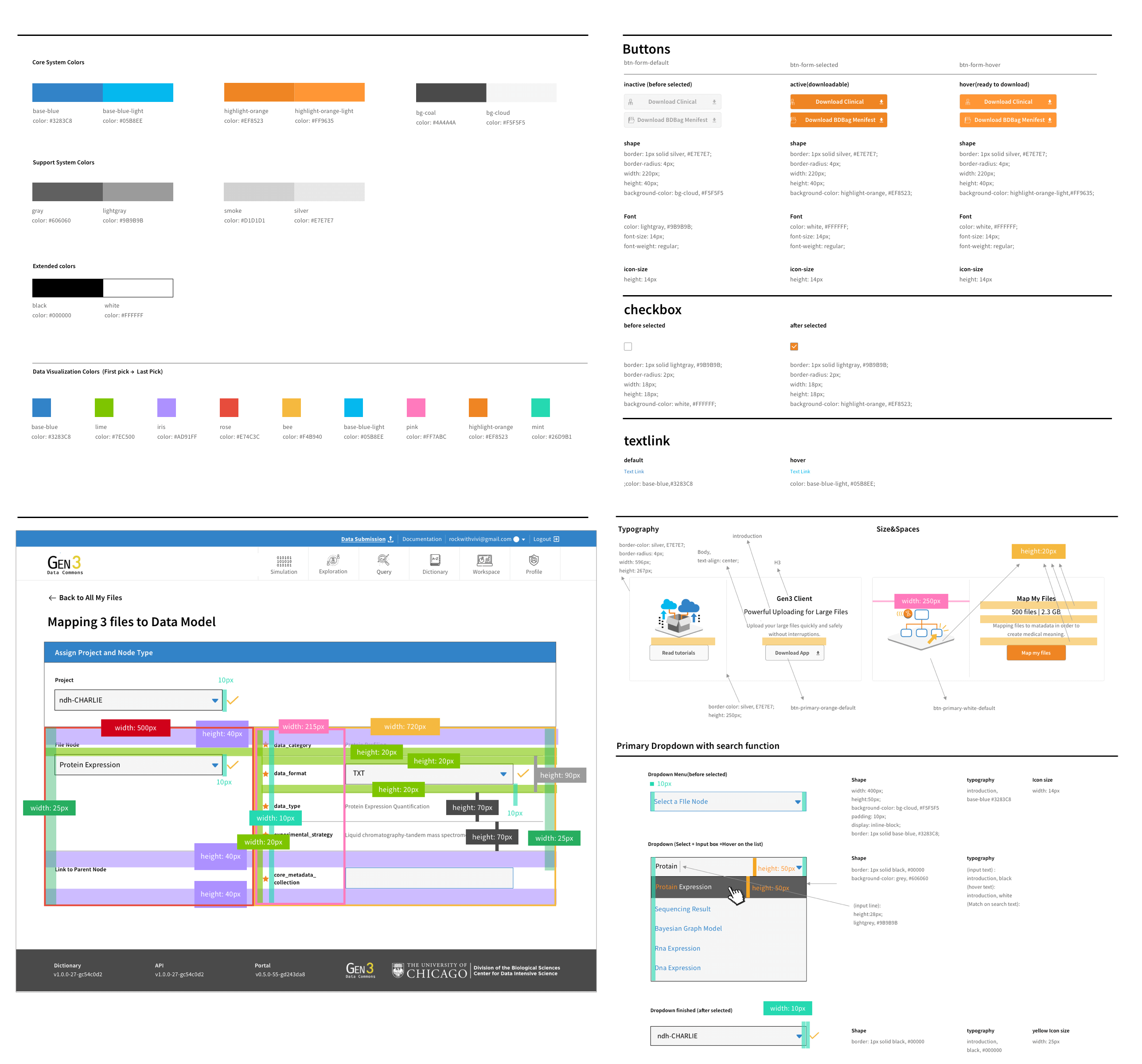
I also extended the design system to incorporate the new design need. I found the process of defining, constructing, and enhancing the atomic design system to be particularly enjoyable.